A glimpse into the gut microbiome of Costa Rican army ants







In the recent paper “Low diversity and host specificity in the gut microbiome community of Eciton army ants (Hymenoptera: Formicidae: Dorylinae) in a Costa Rican rainforest” published in Myrmecological News, Mendoza-Guido and colleagues* investigate the gut microbes of six sympatric Eciton species in a single location in Costa Rica. In the study, the authors used a combination of 16S rRNA amplicon sequencing, fluorescent and electron microscopy to identify and verify gut microbes, and measured the diversity and interaction specificity of the ant-gut microbe interaction network. The results show that the Eciton’s gut microbiome is consistently dominated by a few species of specialized bacteria that may improve nutrient uptake efficiency of host ants. Here, the authors highlight the main points of the study.
* = Bradd Mendoza-Guido, Natalia Rodríguez-Hernández, Aniek Ivens, Christoph Von Beeren, Catalina Murillo-Cruz, Ibrahim Zuniga-Chaves, Piotr Łukasik, Ethel Sanchez, Daniel J. C. Kronauer, and Adrián A. Pinto-Tomás
An Interview compiled by Biplabendu Das, Roberta Gibson, and Patrick Krapf

MNB: Could you tell us a bit about yourself?
BMG: I am a master’s student at the University of Costa Rica and currently work as a molecular biology specialist in a sales company. Since my first years studying biology, I have felt a huge interest in ants and their symbionts, which led me to work on different related projects. Ants are incredible organisms that could teach us a lot about social work.
NRH: I am a biologist and have a master’s degree in microbiology from the University of Costa Rica. Currently, I am working in a medical device company. I am interested in sustainability and environmental topics, that’s why during my master’s I was focused on microbial ecology.
CMC: I am a researcher and professor at the University of Costa Rica. I am also interested in Microbial Ecology.
CVB: I am a postdoc at the Technical University of Darmstadt and a former postdoc at Daniel Kronauer’s Lab in New York. Since my first encounter with Leptogenys army ants in Malaysia in 2007, these remarkable predators have fascinated me. Their role as important arthropod predators and their variety of symbiotic guests are the focus of my research.
IZC: I am a former master’s student at the University of Costa Rica and currently a PhD student at the University of Wisconsin-Madison. I am interested in Microbial Ecology and its applications in human health.
APT: I am a Professor at the University of Costa Rica interested in microbial ecology and symbiosis.
ABFI: I am an evolutionary ecologist who worked for many years on the evolution of mutualism, specifically interactions of ants and other organisms such as aphids and microbes. Like Christoph, I am also a former Postdoc from the Kronauer lab at Rockefeller University. Currently, I am a strategic researcher at a Dutch environmental NGO, and I continue to work on my academic research in my spare time.
MNB: Could you briefly outline the research you published in Myrmecol. News in layperson’s terms?
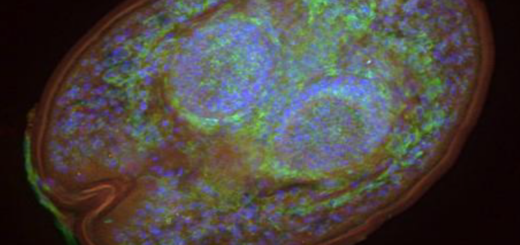
IZC: The army ants are top predators in tropical rainforests. Like most other insects, their gut is colonized by microbes. To learn more about the composition of their gut bacteria, we collected individuals from six closely related species of army ants from a single Costa Rican rainforest. Next, we employed molecular biology to identify the bacteria inside the gut of the collected specimens. Our genetic data shows that the community of microbes in the gut of this group of ants is very simple. It mostly consists of a few bacteria, which we suspect to be bacteria that are beneficial to specifically these ants. We also found that even though we consistently observed the same groups of bacteria over the different species, they did not seem to be specific for each of the six Eciton species. This suggests that these bacteria may be specialized to army ants in general but not specific to any army ant species.
AI: Next to employing genetic techniques to see what bacteria were there, we also employed microscopic techniques to see where the bacteria reside in the ants’ guts. These observations confirmed the findings stated above, confirming that indeed bacteria reside in the ants’ guts and that they may play a role in the ants’ physiology.
IZC: Future work will focus on deciphering the role of these microbes in helping the ants, which we hypothesize could aid in acquiring nutrients or fighting harmful pathogens.
MNB: What is the take-home message of your work?
IZC: The six Eciton army ant species in the same La Selva forest in Costa Rica mostly share the same microbial symbionts, which seem specific to these ants. These microbes might, in fact, represent novel bacterial species.

MNB: What was your motivation for this study?
AAPT: To find out whether army ants receive help from bacteria to maintain their lifestyle, providing services that allow the ants to efficiently digest and obtain energy from their prey.
CvB: My research has mostly focused on arthropod guests of army ants such as beetles, flies, and silverfish. Those macroscopic guests show great variability in host specificity, from species associated with a single Eciton species to generalists that can infiltrate colonies of all locally occurring Eciton species. I was interested in figuring out the level of host specificity in microbic symbionts and how those compare to the macroscopic associations. Interestingly, the macroscopic network shows a higher level of specificity than the microscopic one. Further, I had not worked on microbes yet and thus aimed to learn more about gut microbes – which I did.
ABFI: During my postdoctoral studies, I studied the microbiome in the guts of five ant species (four Lasius species and a Brachymyrmex species) occurring in the same New York forest. These ants have a different lifestyle from army ants: they are not nomadic, and apart from a predatory lifestyle, they also heavily rely on harvesting honeydew for their nutritional needs. I was curious to see how these results would compare to those in army ants. It turns out there are similarities. In the previous system I studied, different ant species share specific species of gut bacteria in the same forest, but there are fewer bacteria species and they are very different from those found in army ants. It turns out there are similarities. For example, in the previous system I studied, different ant species share similar species of gut bacteria within the same forest akin to what we observed in our study, however, they host fewer types of bacteria and they are quite different from those found in army ants.
MNB: What was the biggest obstacle you had to overcome in this project?
AAPT: Completing the publication after the students finished their degrees, which meant re-analyzing the sequence data as platforms and databases are constantly updated.
CvB: I agree with Adrian. I think the main obstacle was the flow of students working on the project, as each student taking over had to familiarize themselves with the work. Further, the cooperation initially started between two institutions – the lab of Daniel at Rockefeller University and the one of Adrian at the University of Costa Rica. After a couple of years, however, several of us moved to different places, had new projects, or even started a job outside of science. Other people with expertise in various topics related to the project joined at some points. I am grateful to the students, namely Naty and Bradd, who did not give up but kept pushing us to finalize the project – thanks for that. And here it is – well done, Naty and Bradd.
ABFI: I couldn’t agree more with Adrian and Christoph. It is quite a feat that Naty and Bradd persisted and finished it! Another obstacle we faced was that this field of microbiome barcoding is developing rapidly. Whenever we picked up the project again, we needed to re-analyze the data according to the latest standards in methodology and technology, for example, the software.
NRH: I agree with Adrian that completing this kind of work is hard after finishing my degree. However, I am glad for the great support of everyone on the team.
MNB: Do you have any tips for others who are interested in doing related research?
AAPT: If available, freezing samples in liquid N2 in the field allows for both DNA-based and culture-based techniques to be applied later.
IZC: It is desirable to combine techniques (for example, DNA sequencing and Fluorescent microscopy) to confirm the presence of potential symbionts in the gut of the insects under study.
CvB: Be sure that you are not allergic to insect stings – being stung from time to time is part of studying army ants.
ABFI: If you do microbiome genotyping and don’t know yet what you’re going to find, it is crucial to work under sterile conditions and run lots of blanks (negative controls). In that way, you will always be able to show that the bacteria you observed were indeed in your original samples.
NRH: Always have a plan B for your samples and an open mind.






Recent Comments