The mystery of mysteries as seen through the morphology of the mound-building red wood ants
In honor of his lifetime work, a commentary by Jonna Kulmuni on Bernhard Seifert’s article “A taxonomic revision of the Palearctic members of the Formica rufa group (Hymenoptera, Formicidae) – the famous mound-building red wood ants” published in Myrmecological News.
A Review by Jonna Kulmuni

While on his voyage on the Beagle, Darwin had little more than his careful observations of morphological characters and distributions of the inhabitants of South America to reach a conclusion on the mystery of mysteries, the origin of species. In his taxonomic revision of the Palearctic Formica rufa group, Seifert uses Numeric Morphology-Based Alpha-Taxonomy (NUMOBAT), by measuring 17 characters in 5500 workers and 24 characters in 410 gynes, to delimit species and clarify the taxonomy of the group. Based on his analysis, Seifert concludes the evolutionary history of the species group is characterized by extreme reticulate evolution and hybridization.
When I started my MSc studies more than 15-years ago, professor Pekka Pamilo presented me with a mystery, a problem, that had bothered him for some years already. It was a population of mound-building red wood ants where he had detected systematic differences between males and gynes with just a few allozyme markers, the first-ever markers used to study genetic differences between species and populations. Genetic differences between sexes at random loci were puzzling because if these ants reproduce sexually, there should be no genetic differences between sexes within a population. Pekka suggested that I have a look at the population with modern genetic markers of that time, the AFLP and microsatellite markers, accompanied by short sequences of the mitochondrial DNA.
I was just as intrigued as Pekka, to find that the males and gynes show genome-wide differentiation within the population! Moreover, we found that the population consisted of two genetic lineages, living side-by-side even in the same nests. The lineages also shared the same mitochondrial haplotype from Formica polyctena. Without any comparison to other ant populations, and based on the genetic data alone, we were unable to say what was happening in the population. That’s when NUMOBAT and Bernhard Seifert entered the picture.
With his extensive measurements, Bernhard was able to show that the workers of the mystery population were morphologically intermediate between two wood ant species, and were actually hybrids between F. polyctena and F. aquilonia. This finding has characterized my whole career. Now, over 15 years later I am still on the same path, trying to understand the causes and consequences of hybridization in the mound-building red wood ants.

In his recent article, published in Myrmecological News, Seifert tackles the taxonomy of the whole Formica rufa group ants and recognizes 13 good species and 32 junior synonyms. A junior synonym in this case refers to a name given for a species that has a name already. The high number of junior synonyms in relation to good species, Seifert writes, reflects the enormous difficulty in interpreting the morphology and phenotypes of these ants. Difficulties arise due to high rates of hybridization, occurrence of local variants, and extreme intraspecific polymorphism.
Thirteen species described by Seifert in his taxonomic revision are F. rufa (Linnaeus, 1761), F. polyctena (Foerster, 1850), F. lugubris (Zetterstedt, 1838), F. helvetica sp.n., F. paralugubris (Seifert, 1996), F. aquilonia (Yarrow, 1955), F. ussuriensis sp.n., F. pratensis (Retzius, 1783), F. kupyanskayae (Bolton, 1995), F. truncorum (Fabricius, 1804), F. dusmeti (Emery, 1909), F. frontalis (Santschi, 1919), and F. sinensis (Wheeler, 1913).
Species delimitation in NUMOBAT is based on measuring 17 morphological characters from several workers per nest with a high-performance stereomicroscope and performing four different analyses using nest centroids as input data. This means the method is primarily based on assigning nests, rather than individuals, into species. The expertise, time, and equipment required for NUMOBAT is something very few scientists have nowadays. And moreover, one might ask, to what extent do measurements of the hairs on the ants reflect species at the genomic level? The short answer is, remarkably well.
I am a person of pragmatism and use the biological species concept where “species are groups of actually or potentially interbreeding natural populations, which are reproductively isolated from other such groups.” As a geneticist, I tend to think the genome is the answer to everything. It reflects the evolutionary history of the species and populations, as well as any reproductive isolation that has accumulated between two or more lineages. However, the reality is not that simple. If speciation happened recently, there may not have been enough time to evolve significant genomic differentiation even if reproductive isolation does exist. Moreover, signatures of reproductive isolation are hard to decipher from other processes that leave their signatures on the genome-wide variation.
Morphology has similar limitations. “NC-clustering of morphological data is no silver bullet to uncover each cryptic species. This became apparent in Formica helvetica sp.n. which was sufficiently separated from Formica lugubris by nuclear DNA. This species is either generally inseparable by worker morphology or the character system applied here does not include the specific diagnostic elements.”, writes Seifert.
Back to the correlation between morphology and the genome. In our recent study (bioRxiv – https://doi.org/10.1101/2021.03.10.434741), we investigate the speciation history of F. aquilonia and F. polyctena using whole-genome sequence data and demographic modeling. Part of the samples used were analyzed with NUMOBAT by Seifert. Species assignment probabilities based on the whole genome data and the morphological assignment based on NUMOBAT are surprisingly similar. Correlation between genetic data from microsatellite markers and the NUMOBAT has also been shown earlier for other species.
Highlighted in several instances of the taxonomic revision are the high rates of hybridization among the Formica rufa group ants. Hybrids can be detected with NUMOBAT, at least to an extent. “Hybridization and introgression were shown or made plausible in 46% of the 13 recognized species with regional hybridization frequencies of above 20% in three species.”, Seifert writes. Moreover, a single species tends to hybridize with multiple other species within the group.
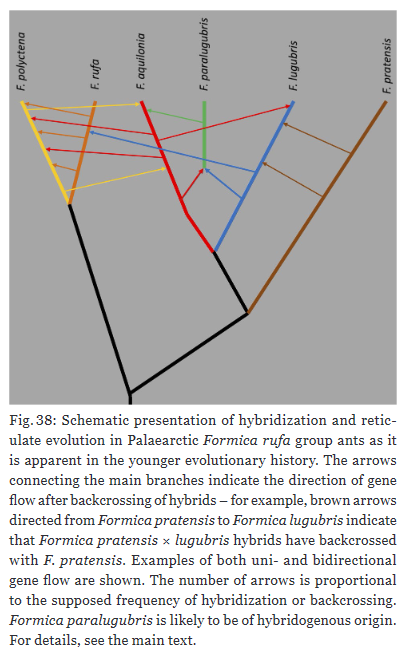
Hybridization is not a mere nuisance in evolution. It is ubiquitous across taxa and a major player in evolution. “Reticulate evolution is a nightmare for taxonomists but an evolutionary biologist’s delight. In the wake of ongoing massive ecosystem change due to human land use and anthropogenic climate warming, repeated introgression of genes may provide an adaptive advantage for long-term survival and prosperity of F. rufa group ants.”, concludes Seifert. With the latter point, I could not agree more.
Ideally, species delimitation is done based on morphological, genomic, behavioral, and ecological data gathered throughout the species’ range. More than a lifetime worth of work for any myrmecologist! With his recent taxonomic revision, Seifert has made a lasting contribution in organizing data and gathering evidence to support his classification of Formica rufa group ants. Now it is up to the rest of us to gather the genomic, behavioral, and ecological data to examine and improve the morphology-based taxonomy of the group.




A very good but a little bit confusing article. But it is from one of the greatest living myrmecologists! More please…